Part:BBa_K2715027
Clostridial promoter Pcdi_tcdB with its native 5' UTR and RBS, driving a GusA reporter
Usage and Biology
This promoter was taken directly from the organism in which we wished to demonstrate our novel methods (C. difficile) for controlling toxin production, as we wanted to gain a greater understanding of the level of expression from this promoter driving expression of tcdB (toxin B) in the wild type organism. For characterisation in C. difficile we decided to use the reporter gene gusA. The GFP protein requires oxygen in order to fluoresce, so it isn’t suitable as a reporter gene in this context. The Β-Glucuronidase encoded by gusA functions as a very sensitive and specific reporter gene in anaerobic organisms. A substrate for the Β-Glucuronidase, 4-methylumbelliferyl glucuronide (4-MUG), can be added to the cell lysate immediately before the assay, and the production of the fluorescent compound 4-methylumbelliferone (4-MU) measured over time in a spectrophotometer. By cloning the native toxin promoter upstream of the gusA reporter gene, we would be able to gain an understanding of whether the toxin genes were actively being expressed under the conditions that we were culturing the organism using a straight forward and highly quantitative assay.
We decided additionally to explore its expression levels in E. coli, as it has been shown in previous research to be under the regulation of an alternative sigma factor encoded by tcdR, present in the C. difficile genome, and therefore we expected to observe reduced expression in E. coli . The variant of this composite part used for testing expression levels in E.coli can be found here BBa_K2715003.
Characterisation
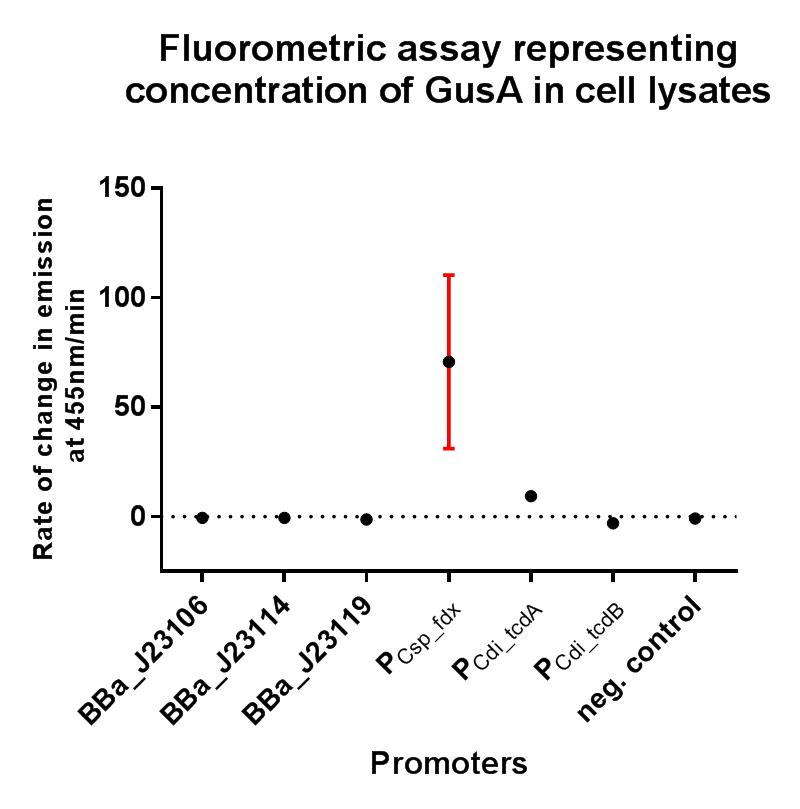
This composite part is composed of the native clostridial promoter driving expression of the toxin gene tcdB, sub-divided into the promoter region itself BBa_K2715013, the 5’ UTR region BBa_K2715023, and the RBS BBa_K2715024, driving expression of gusA taken from K330002.
The gusA containing composites used to assay the promoter activities in C. difficile are listed below.
BBa_K2715025
BBa_K2715026
BBa_K2715027
BBa_K2715028
BBa_K2715029
BBa_K2715030
BBa_K2715031
The plasmid used for this characterisation in C. difficile is displayed below.
Conclusions
This composite part has enabled a standardised characterisation of the native promoter driving the tcdB gene and its strength can be quantified in E. coli using the iGEM 2018 interlab units of fluorescence. It has been demonstrated in this assay to be fairly weakly expressed, suggesting that it may indeed by subject to regulation by an alternative sigma factor, or perhaps it may just be that this clostridial promoter is poorly expressed in the Gram-negative organism E. coli. In the GusA assay the construct BBa_K2715027 showed significantly higher expression of the GusA protein relative to all promoters other than the ferredoxin promoter composite BBa_K2715026. This indicates that this promoter was active and causing toxin expression in our C.difficile cultures, as we have effectively replaced the tcdA gene in C.difficile with the GusA gene and observed expression through measuring fluorescence. Interestingly we did observe expression of the toxin B gene using the composite part BBa_K2715027 in our sample taken during exponential growth, suggesting that these two genes may be subject to slightly different regulation, and validating our experimental approach to targeting both toxin genes separately in order to reduce the toxigenicity of the strains. More time points and further experiments would be useful to investigate the differences in expression of these two promoters.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 139
References
Heap, J.T., Pennington, O.J., Cartman, S.T. and Minton, N.P., 2009. A modular system for Clostridium shuttle plasmids. Journal of microbiological methods, 78(1), pp.79-85.
Davis, D.F., Ward, W.W. and Cutler, M.W., 1994. Posttranslational chromophore formation in recombinant GFP from E. coli requires oxygen. In Bioluminescence and Chemiluminescence: Fundamentals and Applied Aspects. Proceedings of the 8th International Symposium on Bioluminescence and Chemiluminescence, Cambridge. Wiley, New York, NY (pp. 569-599).
Chiu, N.H. and Watson, A.L., 2017. Measuring β‐Galactosidase Activity in Gram‐Positive Bacteria Using a Whole‐Cell Assay with MUG as a Fluorescent Reporter. Current protocols in toxicology, 74(1), pp.4-44.
| None |